Mapping Chromosomes II
What if three traits are linked? We can still calculate map distance, but we can have double crossovers. For instance, a normal fly (+) is crossed with a cis, homozygous recessive fly for three traits: glossy wings (gl), variable sterile (va), and virescent (v). This offspring then produces the gametes:
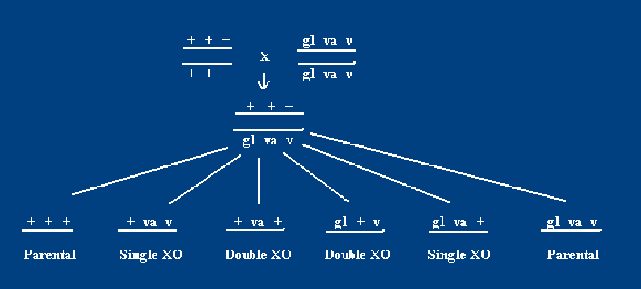
Double crossovers are less frequent than single crossovers.
Let's try an example of multiple trait crossovers.
We crossed a homozygous glossy, virescent fly to a homozygous variable sterile. This offspring will then be crossed to a homozygous recessive fly. What can you conclude from this cross?
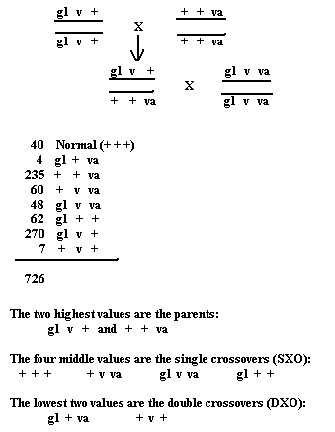
Before distance can be calculated, we must figure the order these genes are located on the chromosome. The best way to do this is to first write down the double crossovers, and then select an order for the parent gametes. Then recheck that to the parents, and when that order fits the double crossover (DXO) pattern, we know that is how they are located on the chromosome.
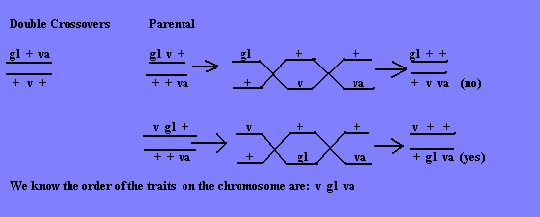
To figure the distance between each trait, we need to first find the distance between the pairs: v-gl and gl-va.
To find the distance of v-gl, add the two single crossovers and the two double crossovers from the parental cross and divide by the total number of offspring.
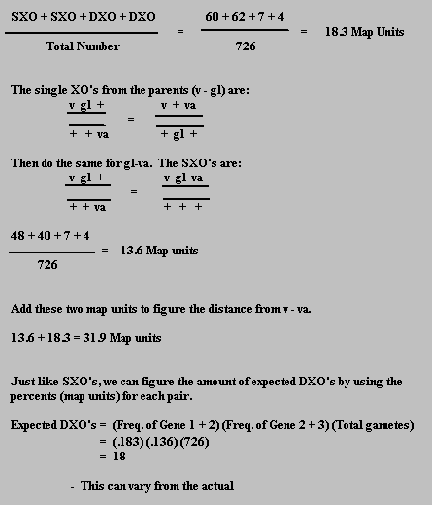
We can also calculate how often we can get the number of expected crossovers by figuring the coefficient of coincidence.
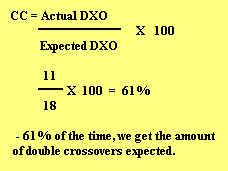
- Continue to Mapping Chromosomes Problem 2
- Return to Main Menu
|
|
© 2003-2026:
USA · USDA · NRPSP8 · Program to Accelerate Animal Genomics Applications.
|
||