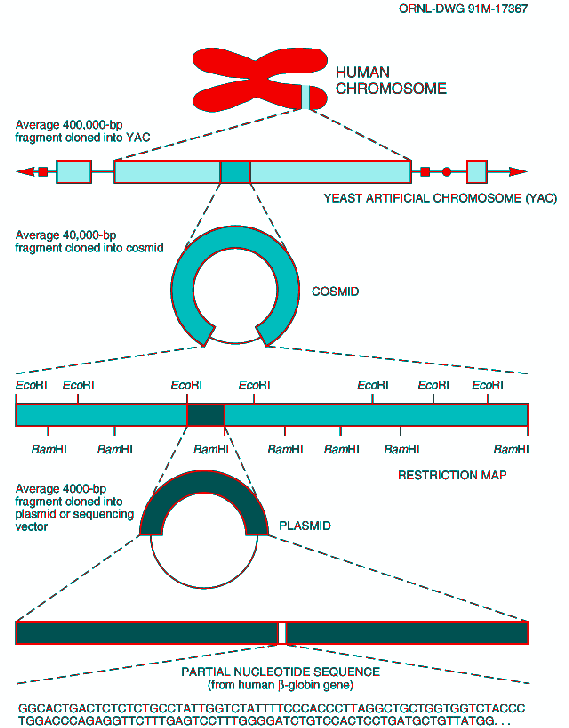
Fig. 11. Constructing Clones for Sequencing.
Cloned DNA molecules must be made progressively smaller and the fragments subcloned into new vectors to obtain fragments small enough for use with current sequencing technology. Sequencing results are compiled to provide longer stretches of sequence across a chromosome. (Source: adapted from David A. Micklos and Greg A. Freyer, DNA Science, A First Course in Recombinant DNA Technology, Burlington, N.C.: Carolina Biological Supply Company, 1990.)
DNA Amplification: Cloning
By fragmenting DNA of any origin (human, animal, or plant) and inserting it in the DNA of rapidly reproducing foreign cells, billions of copies of a single gene or DNA segment can be produced in a very short time. DNA to be cloned is inserted into a plasmid (a small, self- replicating circular molecule of DNA) that is separate from chromosomal DNA. When the recombinant plasmid is introduced into bacteria, the newly inserted segment will be replicated along with the rest of the plasmid.
(b) Constructing an Overlapping Clone Library.
A collection of clones of chromosomal DNA, called a library, has no obvious order indicating the original positions of the cloned pieces on the uncut chromosome. To establish that two particular clones are adjacent to each other in the genome, libraries of clones containing partly overlapping regions must be constructed. These clone libraries are ordered by dividing the inserts into smaller fragments and determining which clones share common DNA sequences.