National Research Support Project- NRSP-8
Year Ending 2002
Craig W. Beattie, Anette Rink
A total of 2,035 ESTs from immune tissues and placentae were used to construct an RH map using the IMpRH7000Rad panel (Yerle et al., 1998; Hawken et al., 1999) and the MAXLIK option of RHMAP V3 (Boehnke et al., 1996) at a 2pt LOD of 6 or 8. The number of observed/expected markers per chromosome ranged from 0.53 for SSC11 to 2.02 for SSC12, respectively. Average marker retention frequency was 30.3%. Annotated ESTs represent 47% of the markers, 30% ESTs showed homology to ESTs from a variety of species, 20% represent novel sequences (ns) with no significant match in any nucleotide or protein database, 3% of the ESTs only identify open reading frames (ORFs) in the human genomic sequence (HGS). More than 28% of the ESTs assigned to the physical map of the pig did not find a homologous sequence in V30 of the human genome assembly. This is indicative of assembly gaps in the HGS.
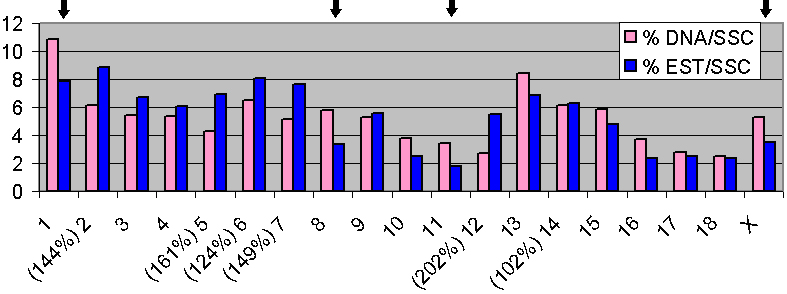
Figure 2. Genome 'Deserts' and Gene Rich Regions in the Porcine Genome. Distribution of 2,035 mapped ESTs. The columns on the left (pink) represent the percentile of DNA in each individual chromosome (Schmitz et al., 1992). Blue columns represent the percentiles of markers mapped. SSC 1, 8, 11 and X (indicated by arrows) correspond to HSA 18, 4, 13 and X, respectively and represent 'gene deserts' both in the human and the porcine genomes. A total of six porcine chromosomes (SSC2, SSC5, SSC6, SSC7, SSC12 and SSC14) are syntenic with the three gene-richest human chromosomes, HSA17, 19 and 22.
The Second Generation EST Radiation Hybrid (RH7000rad) Map (2035 ESTs).
The, as yet unpublished, Second Generation EST Radiation Hybrid Map confirms 98% coverage of the human genome. Currently ~ 60 breaks between chromosomes and ~70 breaks within regions of homology have been detected. A final number cannot be determined at this time since the orientation of a significant number of contigs in HGS Build 29 remains to be determined. Additionally, 4.9% of markers in the current map identify other homologous human chromosomes in HGS Build 29 vs 5.4% in the 1st Generation EST Radiation Hybrid Map. The current porcine RH map extends beyond the genetic linkage map on 38 telomeres. 28.9% of all markers do not find orthologous sequences in the HGS Build 29 vs 23.6% in HGS Build 21, indicating an increasing number of deletions, particularly of telomeric sequences, in subsequent builds of the human genome sequence. The human genome sequence assembly currently does not extend beyond the human RH map(s).
| SSC | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | X |
| Total number of EST markers mapped | 160 | 181 | 138 | 124 | 142 | 165 | 157 | 69 | 114 | 51 | 37 | 112 | 140 | 128 | 98 | 48 | 51 | 48 | 72 |
| Genes | 55 | 74 | 38 | 54 | 63 | 54 | 81 | 27 | 46 | 17 | 11 | 40 | 62 | 52 | 34 | 23 | 20 | 12 | 37 |
| ESTs | 70 | 77 | 72 | 46 | 20 | 82 | 46 | 35 | 49 | 23 | 18 | 53 | 54 | 53 | 43 | 16 | 24 | 18 | 20 |
| novel sequences | 35 | 30 | 28 | 24 | 59 | 29 | 30 | 7 | 19 | 11 | 8 | 19 | 24 | 23 | 21 | 9 | 7 | 18 | 15 |
| Number of linkage groups per chromosome | 15 | 7 | 8 | 5 | 3 | 8 | 5 | 6 | 6 | 3 | 6 | 1 | 12 | 4 | 6 | 5 | 1 | 2 | 7 |
| Number of syntenic chromosomes | 5 | 3 | 3 | 2 | 2 | 4 | 3 | 1 | 3 | 3 | 1 | 1 | 2 | 4 | 2 | 1 | 1 | 1 | 1 |
| ESTs that find matches outside syntenic chromosomes where synteny is based on chromosomal paints | 6 | 9 | 2 | 3 | 5 | 1 | 6 | 5 | 7 | 5 | 3 | 3 | 10 | 5 | 8 | 11 | 8 | 0 | 3 |
| Markers that do not find matches in the HGS V29 | 47 | 59 | 56 | 25 | 31 | 59 | 45 | 18 | 31 | 15 | 12 | 29 | 47 | 38 | 19 | 15 | 9 | 17 | 16 |
| Genes | 2 | 12 | 5 | 5 | 7 | 6 | 12 | 2 | 11 | 1 | 2 | 5 | 7 | 7 | 4 | 3 | 2 | 2 | 2 |
| ESTs | 29 | 24 | 28 | 8 | 14 | 29 | 13 | 12 | 10 | 7 | 5 | 14 | 23 | 14 | 5 | 6 | 5 | 6 | 7 |
| novel sequences | 16 | 23 | 23 | 12 | 10 | 24 | 20 | 4 | 10 | 7 | 5 | 10 | 17 | 17 | 10 | 6 | 2 | 9 | 7 |
We are currently using Carthagene (http://www.inra.fr/bia/T/schiex/Export/WABI.pdf) to calculate an updated version of the 2nd Generation pig human comparative map (IMpRH7000Rad panel). Markers that introduced new homologies to the comparative map are reevaluated based on their multipoint linkage assignment (using the IMpRH mapping tool), rather than on their 2pt LOD value. These maps will be transferred to the FTP site as they become available. An Excel file (Complete Info for 2035 ESTs.xls) containing all pertinent information (annotation, primers, location in HGS build 29, mapping information) is also available from the FTP site. Build 30 of the HGS was released June 24th, 2002 and covers > 70% of the human genome in currently 1,736 contigs assembled, covering 2.87 billion base pairs. BLASTN comparisons against the contig assembly and dbEST have been completed. BLASTP comparisons of the dataset against the CCBC protein database set and nr_NCBI are in progress. Additionally plates MOA131-151 from additional sequencing efforts of the apicomplexin parasite Toxoplasma gondii and its effects on swine epithelial cells, in our laboratory, will be compared against the 1st draft of the Plasmodium falciparum genome as soon as the dataset is available for downloading. An update of both datasets, parasite and (swine) host response genes, is currently under way. We expect to map a significant number of these host response ESTs on the IMpRH12000rad panel.
Radiation Hybrid Mapping project - IMpRH12000rad: We have assigned 102, 138, 124 and 59 ESTs to SSC12, 7, 13 and X, respectively to the IMNpRH212,000-rad panel. These have been finalized and will be available on our web sites (www.toulouse.inra.fr/lgc/lgc/, http://www.ag.unr.edu/ab/standard.htm), shortly. In addition, we have mapped and are finalizing 910 ESTs on SSC1-6 and 68 on SSC8 (current total ~ 1400) that should also be available online shortly, at the above web sites.